* denotes equal contribution; ☨ denotes corresponding author(s)
(Co-) First Author
Expansion in situ genome sequencing links nuclear abnormalities to aberrant chromatin regulation
In this paper, we developed Expansion in situ genome sequencing (ExIGS), a technology that unites 3D DNA sequencing and superresolution imaging in the same cell, which we applied to link nuclear abnormalities to epigenetic dysregulation in aging contexts.
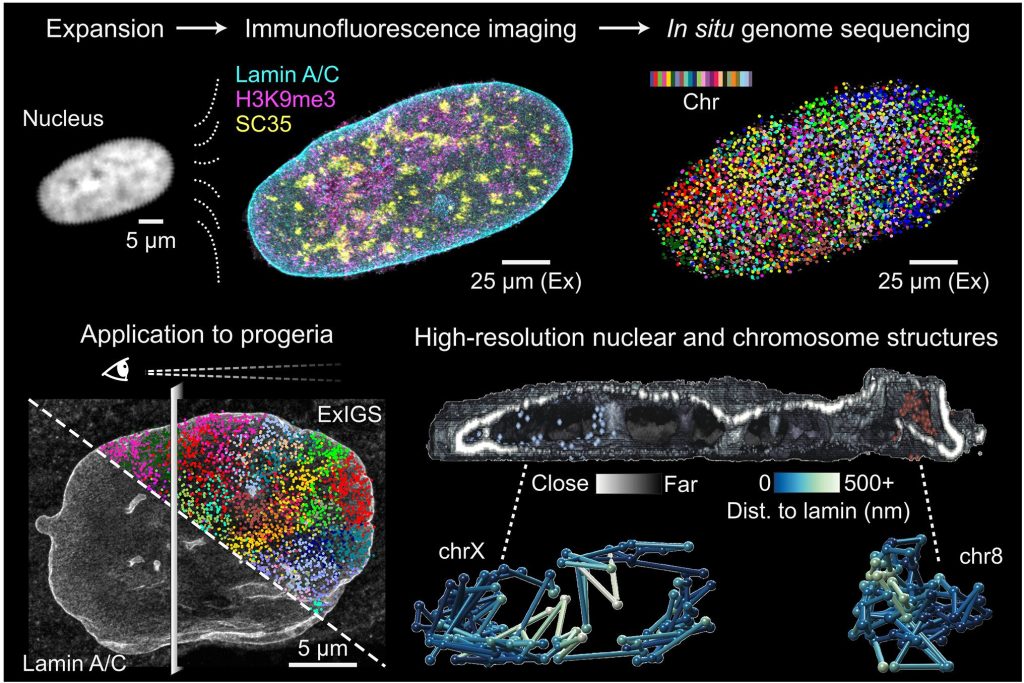
Labade AS*, Chiang ZD*, Comenho C*, Reginato PL, Payne AC, Earl AS, Shrestha R, Duarte FM, Habibi E, Zhang R, Church GM, Boyden ES, Chen F, Buenrostro JD☨. Science (2025).
Featured in Nature and Nature Genetics.
In situ genome sequencing resolves DNA sequence and structure in intact biological samples
In this paper, we developed In situ Genome Sequencing (IGS), a technology that enables 3D DNA sequencing within intact cells, which we applied to examine the first interactions of the maternal and paternal genome in early embryos.
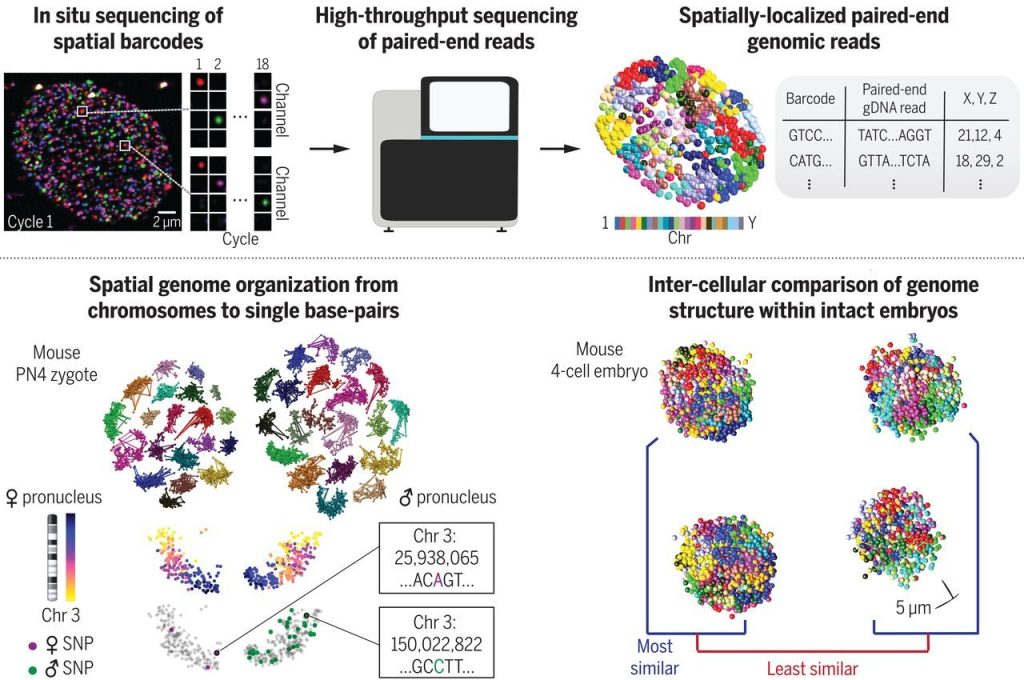
Payne AC*, Chiang ZD*, Reginato PL*, Mangiameli SM, Murray EM, Yao CC, Markoulaki S, Earl AS, Labade AS, Jaenisch R, Church GM, Boyden ES☨, Buenrostro JD☨, Chen F☨. Science (2021).
Featured in Nature Reviews Genetics and Molecular Cell.
Multi-modal spatial genomics enables detection of clonal lineages and their contributions to gene expression
In this paper, we developed Slide-DNA-seq, a method to measure spatial copy number variation in tissues, which we applied to quantify clonal heterogeneity in colon cancer biopsies.
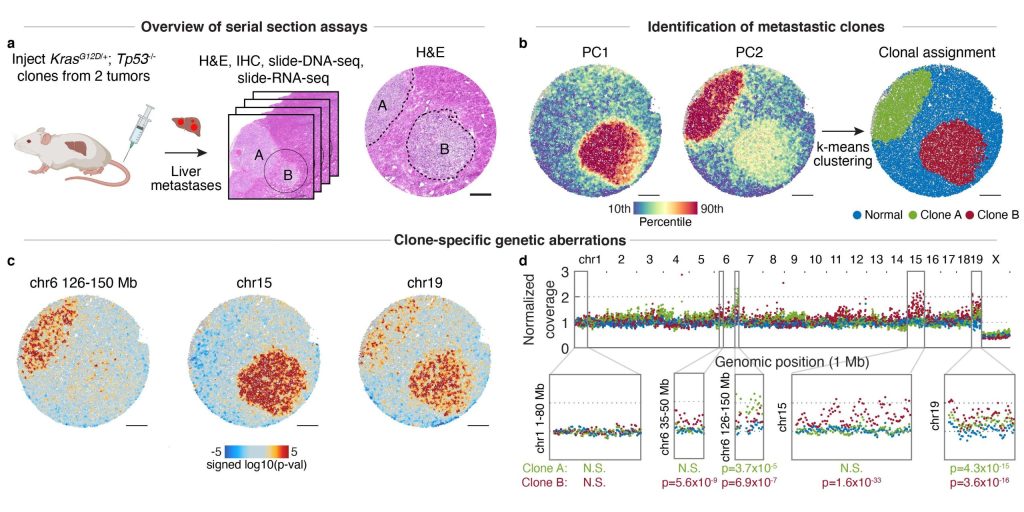
Zhao T*, Chiang ZD*, Morriss JW, LaFave LM, Murray EM, Del Priore I, Meli K, Lareau CA, Nadaf NM, Li J, Macosko EZ, Jacks T, Buenrostro JD☨, Chen F☨. Nature (2022).
Featured in Nature Methods.
Deep learning-based enhancement of epigenomics data with AtacWorks
In this paper, we developed AtacWorks, a deep learning approach for denoising single-cell ATAC-seq data, which we applied to study lineage bias in hematopoietic stem cells.
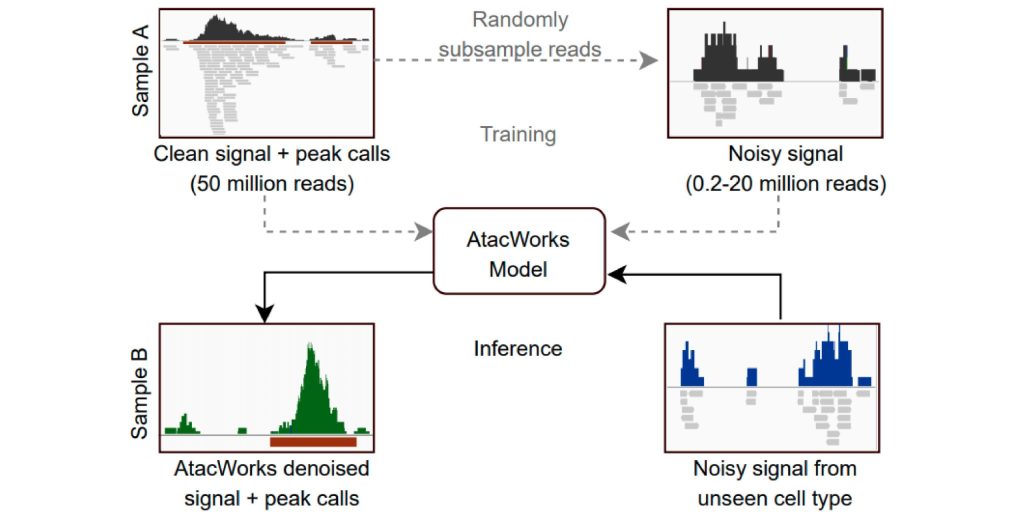
Lal A*, Chiang ZD*, Yakovenko N, Duarte FM, Israeli J☨, Buenrostro JD☨. Nature Communications (2021).
Just happy to be here
Unified molecular approach for spatial epigenome, transcriptome, and cell lineages
Huang YH*, Belk JA*, Zhang R, Weiser NE, Chiang ZD, Jones MG, Mischel PS, Buenrostro JD, Chang H☨. PNAS (2025).
Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling
Lareau CA*, Ludwig LS*, Muus C, Gohil SH, Zhao T, Chiang Z, Pelka K, Verboon JM, Luo W, Christian E, Rosebrock D, Getz G, Boland GM, Chen F, Buenrostro JD, Hacohen N, Wu CJ, Aryee MJ, Regev A☨, Sankaran VJ☨. Nature Biotechnology (2021).
Chromatin potential identified by shared single cell profiling of RNA and chromatin
Ma S, Zhang B, LaFave L, Earl AS, Chiang Z, Hu Y, Ding J, Brack A, Kartha VK, Tay T, Law T, Lareau C, Hsu Y, Regev A☨, Buenrostro JD☨. Cell (2020).
Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma
LaFave LM, Kartha VK*, Ma S*, Meli K, Priore ID, Lareau C, Naranjo S, Westcott P, Duarte FM, Sankar V, Chiang Z, Brack A, Law T, Hauck H, Okimoto A, Regev A, Buenrostro JD☨, Tyler Jacks☨. Cancer Cell (2020).
A compendium of promoter-centered long-range chromatin interactions in the human genome
Jung I*, Schmitt A*, Diao Y*, Lee AJ, Liu T, Yang D, Tan C, Eom J, Chan M, Chee S, Chiang Z, Kim C, Masliah E, Barr CL, Li B, Kuan S, Kim D, Ren B☨. Nature Genetics (2019).